Abstract
Immunomodulatory drugs (IMiDs) and proteasome inhibitors (PI) are backbone agents in the treatment of Multiple Myeloma (MM). However, most patients develop drug resistance over time, with the underlying mechanisms poorly understood. Epigenetic modifications are effective modulatory mechanisms for gene silencing and known to be influenced by the exposure to environmental factors such as drug treatment. Dimopoulous et al. (Molecular Oncology, 2018) recently demonstrated that resistance to IMiDs is also associated with global changes in DNA methylation, although such changes were absent in the annotated promoter region of the CRBN gene. Similarly, DNA methylation of PSMD5 and other regulatory 19S proteasome subunit genes induces PI resistance in various cancer cell lines (Tsvetkov et al. PNAS 2017, eLive 2015).
Deep bisulfite sequencing (DBS) is a targeted sequencing approach that allows to investigate CpG methylation at single DNA molecule resolution. In order to determine the role of promoter methylation in the development of drug resistance in MM, we analyzed primary tumor samples for a selected number of therapy-associated genes, i.eCRBN, PSMD5, TP53, MDM2 and cMYC. A total of 59 sequential time points (TP, CD138 purified MM cells) from 32 MM and 30 paired PBMCs were investigated. At least one TP for all the patients was in relapse to IMiDs and/or PIs. Five samples of (un)methylated DNA standards (0%, 25%, 50%, 75%, and 100%) were also included for every gene in order to confirm assay specificity. DBS primers for multiplexed libraries were designed using the PyroMark Assay Design 2.0 software (Qiagen). Per sample, 200ng of genomic DNA was bisulfite converted using the EpiTect® Fast 96 Bisulfite Conversion Kit (Qiagen) and the DBS libraries were sequenced with the Illumina MiSeq. FASTQ files were analyzed using the Amplikyzer2 pipeline. The median read coverage per sample and gene was 10,031X ± 6624 (SD). A locus was considered to be affected when the average methylation was higher than 15%.
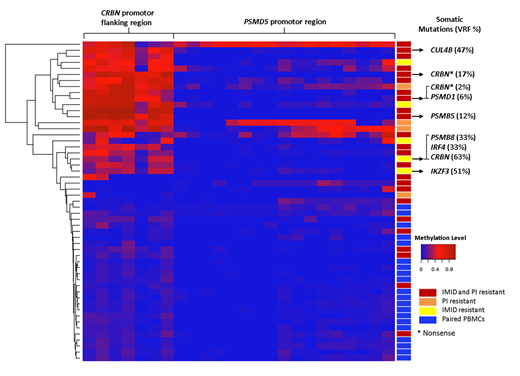
We confirmed the absence of methylation in the CRBN promoter region, as previously described, and found an unmethylated pattern in all analyzed samples (MM and PBMCs) for the tumor suppressor gene TP53 and the oncogenes MDM2 and cMYC. Strikingly, the CRBN promoter flanking region which lays 6,039bp downstream of the promoter, was identified to be highly methylated. 18 out of 28 IMiD resistant patients (64%) showed a hypermethylated state in this region (average methylation: 48.7%, range: 22.7%-83.3%), whereas none of the corresponding PBMCs was affected (5.7%; range, 2.6%-8.8%). Similarly, but to a lesser extent, PSMD5 hypermethylation was observed in 4 out of 27 (15%) PI resistant patients affected (MM cells; 31.3%, range 22.4%-50.8%, PMBCs; 2.0%, range; 0.8%-3.4%).
Notably, in six patients that were previously exposed to IMiDs and PIs, an aberrant co-methylation of CRBN and PSMD5 was detected, with four of them being clearly resistant to both drug families.
In three patients with longitudinal sampling, one was unmethylated at diagnosis in both genes. In PI- and IMiD-resistant relapse of the same patient four years later, methylation was detected in 27% of PSMD5 reads and 62% of the CRBN flanking region. The second patient showed 70% methylation in CRBN at primary diagnosis and 83% in IMiD-resistant relapse six years later, indicating a pre-existing condition. Of note this patient did not respond to Lenalidomide induction therapy. The third patient had no detectable methylation at diagnosis and relapse. In the remaining 12 patients with sequential sampling, we identified highly conserved methylation patterns in both genes, indicating certain stability of methylation pattern once selected. In addition, six patients with mutations in proteasome subunits and/or the CRL4/CRBN complex also showed methylation in CRBN or PSMD5. This proves that genomic mutations and aberrant methylation patterns are not mutually exclusive.
Here we provide first evidence that the methylation of cancer driver genes like TP53 or cMYC, seem to be strictly regulated, whereas genes targeted by therapy like CRBN or PSMD5 have a dynamic methylation pattern. The accumulation of epimutations as a potential mechanism for drug resistance needs to be confirmed in larger cohorts, a recovering of the gene expression might provide a valid treatment strategy to explore.
Martinez Lopez:Celgene: Research Funding, Speakers Bureau; Bristol Myers Squibb: Research Funding, Speakers Bureau; Janssen: Research Funding, Speakers Bureau; Novartis: Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal